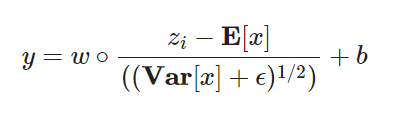cmu 10-414 HW2 Question 2 1 2 3 4 5 def xavier_uniform (fan_in, fan_out, gain=1.0 , **kwargs ): a = gain * math.sqrt(6 / (fan_in + fan_out)) out = rand(fan_in, fan_out, low=-a, high=a) return out
Xavier normal 1 2 3 4 5 def xavier_normal (fan_in, fan_out, gain=1.0 , **kwargs ): std = gain * math.sqrt(2 / (fan_in + fan_out)) out = randn(fan_in, fan_out, mean=0 , std=std) return out
1 2 3 4 5 6 7 def kaiming_uniform (fan_in, fan_out, nonlinearity="relu" , **kwargs ): assert nonlinearity == "relu" , "Only relu supported currently" gain = math.sqrt(2 ) bound = gain * math.sqrt(3 / fan_in) out = rand(fan_in, fan_out, low=-bound, high=bound) return out
Kaiming normal 1 2 3 4 5 6 def kaiming_normal (fan_in, fan_out, nonlinearity="relu" , **kwargs ): assert nonlinearity == "relu" , "Only relu supported currently" gain = math.sqrt(2 ) std = gain / math.sqrt(fan_in) out = randn(fan_in, fan_out, mean=0 , std=std) return out
Question 2 Linear 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 class Linear (Module ): def __init__ ( self, in_features, out_features, bias=True , device=None , dtype="float32" ): super ().__init__() self.in_features = in_features self.out_features = out_features self.have_bias = bias self.weight = Parameter(init.kaiming_uniform(in_features, out_features,device = device, dtype = dtype)) if self.have_bias: self.bias = Parameter(init.kaiming_uniform(out_features, 1 , device = device, dtype = dtype).reshape((1 , out_features))) def forward (self, X: Tensor ) -> Tensor: if self.have_bias: bias = ops.broadcast_to(self.bias, (X.shape[0 ], self.out_features)) return ops.matmul(X, self.weight) + bias else : return ops.matmul(X, self.weight)
Sequential 1 2 3 4 5 6 7 8 9 10 11 class Sequential (Module ): def __init__ (self, *modules ): super ().__init__() self.modules = modules def forward (self, x: Tensor ) -> Tensor: input = x for module in self.modules: input = module(input ) return input
LogSumExp 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 class LogSumExp (TensorOp ): def __init__ (self, axes: Optional [tuple ] = None ): self.axes = axes def compute (self, Z ): max_z_f = array_api.max (Z, axis=self.axes, keepdims=False ) max_z_t = array_api.max (Z, axis=self.axes, keepdims=True ) tmp = array_api.sum (array_api.exp(Z - max_z_t),axis=self.axes, keepdims=False ) out = array_api.log(tmp) + max_z_f return out def gradient (self, out_grad, node ): input = node.inputs[0 ].cached_data max_val_t = array_api.max (input , axis=self.axes, keepdims=True ) exp_val = array_api.exp(input - max_val_t) sum_val = array_api.sum (exp_val, axis=self.axes, keepdims=False ) grad_log = out_grad.cached_data / sum_val shape = list (node.inputs[0 ].shape) axes = self.axes if axes is None : axes = list (range (len (shape))) for _ in axes: shape[_] = 1 grad_sum = array_api.broadcast_to(array_api.reshape(grad_log, shape), node.inputs[0 ].shape) grad_exp = grad_sum * exp_val return Tensor(grad_exp)
SoftmaxLoss 1 2 3 4 5 6 7 8 9 10 11 class SoftmaxLoss (Module ): def forward (self, logits: Tensor, y: Tensor ): softmax = ops.logsumexp(logits, axes=(1 ,)) shape = logits.shape batch_size = shape[0 ] num_class = shape[1 ] y_one_hot = init.one_hot(num_class, y) I = ops.summation(logits * y_one_hot, axes=(1 ,)) return ops.summation(softmax - I) / batch_size
LayerNorm1d 为什么需要Normalization?
深度神经网络模型的训练为什么会很困难?其中一个重要的原因是,深度神经网络涉及到很多层的叠加,而每一层的参数更新会导致上层的输入数据分布发生变化,通过层层叠加,高层的输入分布变化会非常剧烈,这就使得高层需要不断去重新适应底层的参数更新。为了训好模型,我们需要非常谨慎地去设定学习率、初始化权重、以及尽可能细致的参数更新策略。Google 将这一现象总结为 Internal Covariate Shift,简称 ICS.
大家都知道在统计机器学习中的一个经典假设是“源空间(source domain)和目标空间(target domain)的数据分布(distribution)是一致的”。如果不一致,那么就出现了新的机器学习问题,如 transfer learning / domain adaptation 等。而 covariate shift 就是分布不一致假设之下的一个分支问题,它是指源空间和目标空间的条件概率是一致的,但是其边缘概率不同,即:对所有𝑥∈𝑋,𝑃𝑠(𝑌|𝑋=𝑥)=𝑃𝑡(𝑌|𝑋=𝑥) 但是𝑃𝑠(𝑋)≠𝑃𝑡(𝑋) 大家细想便会发现,的确,对于神经网络的各层输出,由于它们经过了层内操作作用,其分布显然与各层对应的输入信号分布不同,而且差异会随着网络深度增大而增大,可是它们所能“指示”的样本标记(label)仍然是不变的,这便符合了covariate shift的定义。由于是对层间信号的分析,也即是“internal”的来由。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 """ 开始时计算E时,想直接使用.data来计算,不想创建复杂的计算图。后来意识到这计算过程就是需要创建计算图的。乐 """ class LayerNorm1d (Module ): def __init__ (self, dim, eps=1e-5 , device=None , dtype="float32" ): super ().__init__() self.dim = dim self.eps = eps self.weight = Parameter(init.ones(dim, device=device, dtype=dtype)) self.bias = Parameter(init.zeros(dim, device=device, dtype=dtype)) def forward (self, x: Tensor ) -> Tensor: batch_size = x.shape[0 ] features = x.shape[1 ] sum_x = ops.summation(x, 1 ) mean_x = ops.divide_scalar(sum_x, features) mean_x_reshape = ops.reshape(mean_x, (-1 ,1 )) E = ops.broadcast_to(mean_x_reshape, x.shape) V_inner = ops.power_scalar(x-E, 2 ) V_sum = ops.summation(V_inner, 1 ) V_mean = ops.divide_scalar(V_sum, features) V = ops.add_scalar(V_mean, self.eps) sqrt_Var = ops.power_scalar(V, 1 /2 ) sqrt_Var_reshape = ops.reshape(sqrt_Var, (-1 ,1 )) sqrt_Var_reshape_brocst = ops.broadcast_to(sqrt_Var_reshape, x.shape) X = (x - E) / sqrt_Var_reshape_brocst broadcast_weight = ops.broadcast_to(ops.reshape(self.weight, (1 , -1 )), x.shape) broadcast_bias = ops.broadcast_to(ops.reshape(self.bias, (1 ,-1 )), x.shape) out = broadcast_weight * X + broadcast_bias return out
BatchNorm1d
LayerNormalize是在一个sample中对所有dimension进行的。而BatchNormalize是针对一个dim将minibatch中该dim所有的值进行规范
公式分两步:
中间的计算使得第 𝑙 层的输入每个特征的分布均值为0,方差为1。
引入可学习的参数w和b,引入的目的是为了恢复数据本身的表达能力,对规范化后的数据进行线性变换
在训练模型时,在每一层计算的 𝜇 与 𝜎2 都是基于当前batch中的训练数据;而当做预测时,一批预测只有一个或者很少样本,计算出的𝜇 与 𝜎2 一定有偏差。如何解决?
保留每组mini-batch训练数据在网络中每一层的 𝜇𝑏𝑎𝑡𝑐ℎ 与 𝜎𝑏𝑎𝑡𝑐ℎ2 。预测时使用整个样本的统计量来对预测数据进行归一化。即额外计算每层网络的运行时均值和方差
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 class BatchNorm1d (Module ): def __init__ (self, dim, eps=1e-5 , momentum=0.1 , device=None , dtype="float32" ): super ().__init__() self.dim = dim self.eps = eps self.momentum = momentum self.weight = Parameter(init.ones(dim, device=device, dtype=dtype)) self.bias = Parameter(init.zeros(dim, device=device, dtype=dtype)) """ 下边两个并不是Parameter,因为他们不是权重,在最后一个测试时发现 """ self.running_mean = init.zeros(dim, device=device, dtype=dtype) self.running_var = init.ones(dim, device=device, dtype=dtype) def forward (self, x: Tensor ) -> Tensor: batch_size = x.shape[0 ] broadcast_weight = ops.broadcast_to(ops.reshape(self.weight, (1 , -1 )), x.shape) broadcast_bias = ops.broadcast_to(ops.reshape(self.bias, (1 ,-1 )), x.shape) if self.training: sum_x = ops.summation(x, 0 ) mean_x = ops.divide_scalar(sum_x, batch_size) broadcast_mean = ops.broadcast_to(mean_x, x.shape) pow_Var = ops.power_scalar(x-broadcast_mean, 2 ) sum_Var = ops.summation(pow_Var, 0 ) Var = ops.divide_scalar(sum_Var, batch_size) Var_add_eps = ops.power_scalar(ops.add_scalar(Var, self.eps), 1 /2 ) broadcast_var = ops.broadcast_to(Var_add_eps, x.shape) out = broadcast_weight * (x - broadcast_mean) / broadcast_var + broadcast_bias self.running_mean = (1 - self.momentum) * self.running_mean + self.momentum * mean_x self.running_var = (1 - self.momentum) * self.running_var + self.momentum * Var else : broadcast_running_mean = ops.broadcast_to(ops.reshape(self.running_mean, (1 ,-1 )), x.shape) running_var_add_eps = ops.power_scalar(ops.add_scalar(self.running_var, self.eps), 1 /2 ) broadcast_running_var = ops.broadcast_to(ops.reshape(running_var_add_eps, (1 ,-1 )), x.shape) out = broadcast_weight * (x-broadcast_running_mean) / broadcast_running_var + broadcast_bias return out
Dropout 1 2 3 4 5 6 7 8 9 10 class Dropout (Module ): def __init__ (self, p=0.5 ): super ().__init__() self.p = p def forward (self, x: Tensor ) -> Tensor: if self.training: mask = init.randb(*x.shape, p = 1 - self.p) / (1 - self.p) x = x * mask return x
Residual 1 2 3 4 5 6 7 class Residual (Module ): def __init__ (self, fn: Module ): super ().__init__() self.fn = fn def forward (self, x: Tensor ) -> Tensor: return self.fn(x) + x
Question3 SGD 有几个需要注意的
u是字典,key是Param,value是该Param上一次计算时的梯度。
weight_decay表明需要考虑L2regularization,即真正的梯度是param.grad.data + weight_decay * W,这才是(1-β)后的那个梯度
因为写完后报错float64和float32不一致,所以将grad的dtype改成param的dtype
1 2 3 4 5 6 7 8 9 10 11 12 13 14 class SGD (Optimizer ): def __init__ (self, params, lr=0.01 , momentum=0.0 , weight_decay=0.0 ): super ().__init__(params) self.lr = lr self.momentum = momentum self.u = {} self.weight_decay = weight_decay def step (self ): for param in self.params: grad = self.u.get(param, 0 ) * self.momentum + (1 -self.momentum) * (param.grad.data + self.weight_decay * param.data) grad = ndl.Tensor(grad, dtype = param.dtype) self.u[param] = grad param.data = param.data - self.lr * grad
Adam 增加 if param.grad is not None:判断条件是因为报错param.grad是NoneType的情况
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 class Adam (Optimizer ): def __init__ ( self, params, lr=0.01 , beta1=0.9 , beta2=0.999 , eps=1e-8 , weight_decay=0.0 , ): super ().__init__(params) self.lr = lr self.beta1 = beta1 self.beta2 = beta2 self.eps = eps self.weight_decay = weight_decay self.t = 0 self.m = {} self.v = {} def step (self ): self.t += 1 for param in self.params: if param.grad is not None : grad_with_L2rgl = param.grad.data + self.weight_decay * param.data else : grad_with_L2rgl = self.weight_decay * param.data new_m = self.beta1 * self.m.get(param, 0 ) + (1 -self.beta1) * grad_with_L2rgl.data new_v = self.beta2 * self.v.get(param, 0 ) + (1 -self.beta2) * grad_with_L2rgl.data * grad_with_L2rgl.data self.m[param] = new_m self.v[param] = new_v m_hat = new_m.data / (1 - self.beta1 ** self.t) v_hat = new_v.data / (1 - self.beta2 ** self.t) out = param.data - self.lr * m_hat / (ndl.ops.power_scalar(v_hat, 1 /2 ) + self.eps) out = ndl.Tensor(out, dtype=param.dtype) param.data = out
Question4 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 class RandomFlipHorizontal (Transform ): def __init__ (self, p = 0.5 ): self.p = p def __call__ (self, img ): """ Horizonally flip an image, specified as an H x W x C NDArray. Args: img: H x W x C NDArray of an image Returns: H x W x C ndarray corresponding to image flipped with probability self.p Note: use the provided code to provide randomness, for easier testing """ flip_img = np.random.rand() < self.p if flip_img: img = img[:,::-1 ,:] return img
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 class RandomCrop (Transform ): def __init__ (self, padding=3 ): self.padding = padding def __call__ (self, img ): """ Zero pad and then randomly crop an image. Args: img: H x W x C NDArray of an image Return H x W x C NAArray of cliped image Note: generate the image shifted by shift_x, shift_y specified below """ shift_x, shift_y = np.random.randint(low=-self.padding, high=self.padding+1 , size=2 ) H, W, C = img.shape pad = np.zeros((H+2 *self.padding, W+2 *self.padding, C)) pad[self.padding:self.padding+H,self.padding:self.padding+W,:] = img x = shift_x + self.padding y = shift_y + self.padding img = pad[x:x+H, y:y+W,:] return img
MNISTDataset Dataset的作用是对于给定的数据路径,读取其中的数据
parse_mnist是从hw0复制来的,但是由于上边的转换函数我们发现对于一个sample是H* W* C,因此输出的X的维度应该是(sample_numbles, H, W, C)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 class MNISTDataset (Dataset ): def __init__ ( self, image_filename: str , label_filename: str , transforms: Optional [List ] = None , ): self.images, self.labels = parse_mnist(image_filename, label_filename) self.transforms = transforms def __getitem__ (self, index ) -> object : img = self.images[index] return self.apply_transforms(img), self.labels[index] def __len__ (self ) -> int : return self.images.shape[0 ] def parse_mnist (image_filename, label_filename ): with gzip.open (image_filename, 'rb' ) as img_f: img_f.read(4 ) num_images = int .from_bytes(img_f.read(4 ), 'big' ) rows = int .from_bytes(img_f.read(4 ), 'big' ) cols = int .from_bytes(img_f.read(4 ), 'big' ) image_data = img_f.read(num_images * rows * cols) X = np.frombuffer(image_data, dtype=np.uint8).astype(np.float32) X = X.reshape(num_images, rows, cols, 1 ) X /= 255.0 with gzip.open (label_filename, 'rb' ) as lb_f: lb_f.read(4 ) num_labels = int .from_bytes(lb_f.read(4 ), 'big' ) lable_data = lb_f.read(num_labels) y = np.frombuffer(lable_data, dtype=np.uint8) return X, y
Dataloader 给定Dataset,将数据按batch_size进行分批,并通过iter和next实现迭代。每次训练输入一批数据。需要注意一个坑:shuffle表示数据集中的sample是否需要打乱。开始时我直接在init()函数中判断,如果为true,直接做数据的打乱,而在iter函数中直接return self代码如下
1 2 3 4 5 6 7 8 9 if not self.shuffle: self.ordering = np.array_split(np.arange(len (dataset)), range (batch_size, len (dataset), batch_size)) else : self.ordering = np.array_split(np.random.permutation(len (self.dataset)), range (self.batch_size, len (self.dataset), self.batch_size)) def __iter__ (self ): return self
而这么做的问题是,一旦在init中打乱,那么当做多epoch的训练时,对于不同的epoch来说,其ordering中的minibatch是一样的,因为只打乱了一次。
因此不能再init中打乱,而需要在iter中打乱。这样在每个epoch中,对minibatch的迭代开始时调用一次iter,到下一个epoch时,对minibatch迭代会再次调用iter,保证不同epoch中minibatch都不同
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 class DataLoader : r""" Data loader. Combines a dataset and a sampler, and provides an iterable over the given dataset. Args: dataset (Dataset): dataset from which to load the data. batch_size (int, optional): how many samples per batch to load (default: ``1``). shuffle (bool, optional): set to ``True`` to have the data reshuffled at every epoch (default: ``False``). """ dataset: Dataset batch_size: Optional [int ] def __init__ ( self, dataset: Dataset, batch_size: Optional [int ] = 1 , shuffle: bool = False , ): self.dataset = dataset self.shuffle = shuffle self.batch_size = batch_size if not self.shuffle: self.ordering = np.array_split(np.arange(len (dataset)), range (batch_size, len (dataset), batch_size)) def __iter__ (self ): if self.shuffle: self.ordering = np.array_split(np.random.permutation(len (self.dataset)), range (self.batch_size, len (self.dataset), self.batch_size)) self.idx = -1 return self def __next__ (self ): self.idx += 1 if self.idx >= len (self.ordering): self.idx = -1 raise StopIteration() samples = self.dataset[self.ordering[self.idx]] samples = [Tensor(s) for s in samples] return samples
Question 5 ResidualBlock 1 2 3 4 5 6 7 8 9 10 def ResidualBlock (dim, hidden_dim, norm=nn.BatchNorm1d, drop_prob=0.1 ): main = nn.Sequential( nn.Linear(dim, hidden_dim), norm(hidden_dim), nn.ReLU(), nn.Dropout(drop_prob), nn.Linear(hidden_dim, dim), norm(dim), ) return nn.Sequential(nn.Residual(main), nn.ReLU())
MLPResNet 作业的图中是没用Flatten的,但是输入的X的dim是(batch_size, H, W, C), 而第一个W的维度是(dim, hidden_dim),这里的dim=H* W* 1。因此要给X进行Flatten
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 def MLPResNet ( dim, hidden_dim=100 , num_blocks=3 , num_classes=10 , norm=nn.BatchNorm1d, drop_prob=0.1 , layers = [] layers.append(nn.Flatten()) layers.append(nn.Linear(dim, hidden_dim)) layers.append(nn.ReLU()) for i in range (num_blocks): layers.append(ResidualBlock(hidden_dim, hidden_dim // 2 , norm, drop_prob)) layers.append(nn.Linear(hidden_dim, num_classes)) return nn.Sequential(*layers)
Epoch 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 def epoch (dataloader, model, opt=None ): np.random.seed(4 ) if opt is None : model.eval () else : model.train() loss_fuc = nn.SoftmaxLoss() acc_num = 0 losses = [] for X, y in dataloader: out = model(X) loss = loss_fuc(out, y) if opt is not None : loss.backward() opt.step() losses.append(loss.numpy()) acc_num += (out.numpy().argmax(axis=1 ) == y.numpy()).sum () return 1 - acc_num / len (dataloader.dataset), np.mean(losses)
Train Mnist 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 def train_mnist ( batch_size=100 , epochs=10 , optimizer=ndl.optim.Adam, lr=0.001 , weight_decay=0.001 , hidden_dim=100 , data_dir="data" , np.random.seed(4 ) trainning_dataset = ndl.data.MNISTDataset( os.path.join(data_dir, "train-images-idx3-ubyte.gz" ), os.path.join(data_dir, "train-labels-idx1-ubyte.gz" ) ) trainning_data_loader = ndl.data.DataLoader(trainning_dataset, batch_size, shuffle=True ) test_dataset = ndl.data.MNISTDataset( os.path.join(data_dir, "t10k-images-idx3-ubyte.gz" ), os.path.join(data_dir, "t10k-labels-idx1-ubyte.gz" ) ) test_data_loader = ndl.data.DataLoader(test_dataset, batch_size) shape = test_data_loader.dataset.images.shape dim = shape[1 ] * shape[2 ] print (dim) model = MLPResNet(dim=dim, hidden_dim=hidden_dim) opt = optimizer(model.parameters(), lr=lr, weight_decay=weight_decay) train_err,train_loss = 0 , 0 test_err,test_loss = 0 , 0 for i in range (epochs): train_err, train_loss = epoch(trainning_data_loader, model, opt) print ("Epoch %d: Train err: %f, Train loss: %f" % ( i, train_err, train_loss )) test_err, test_loss = epoch(test_data_loader, model) return train_err, train_loss, test_err, test_loss